The Nephrotic Syndrome Study Network (NEPTUNE) is a collaborative investigational infrastructure of 23 sites across North America for conducting clinical and translational research on a group of rare kidney diseases presenting as Nephrotic Syndrome: Focal and Segmental Glomerular Sclerosis (FSGS), Minimal Change Disease (MCD), and Membranous Nephropathy (MN). Currently, the clinical classification of nephrotic syndrome relies primarily on descriptive conventional histopathology. From a clinical perspective, histopathology alone does not adequately predict the heterogeneous natural history or response to therapy of individuals within a given glomerular disease diagnosis 1, 2. This clinical heterogeneity might be caused by distinct molecular mechanisms that each present with clinically indistinguishable histopathology. Given our poor understanding of MCD, FSGS, and MN pathobiology and our inadequate and misleading clinical classification system, it is not surprising that our therapeutic approach to these diseases is also imperfect. Current therapies rely almost exclusively on immunosuppression, which is used without a clear mechanistic basis, is often not beneficial, and frequently complicated by significant toxicities.
NEPTUNE has embraced the concept of Precision Medicine proposed by the Institute of Medicine in 2011 3 (Fig. 1). Charged by the National Research Council of the National Academies to identify a strategy towards a “New Taxonomy of human disease based on molecular biology”, the Precision Medicine strategy proposes the development of new disease definitions generated by a multilayered analysis of the disease course while patients receive their usual clinical care in observational studies. These studies provide comprehensive data sets along the genotype – phenotype continuum into an “Informational Commons” that basic and clinical scientists use to develop a “Knowledge Network” driving disease classification from a molecular understanding of the phenotypic features captured in the cohorts. The new, molecular ‘Taxonomy’ allows for more accurate diagnosis and targeted treatment to improve health outcomes (Fig.1). From inception in 2009, NEPTUNE has adopted this model.
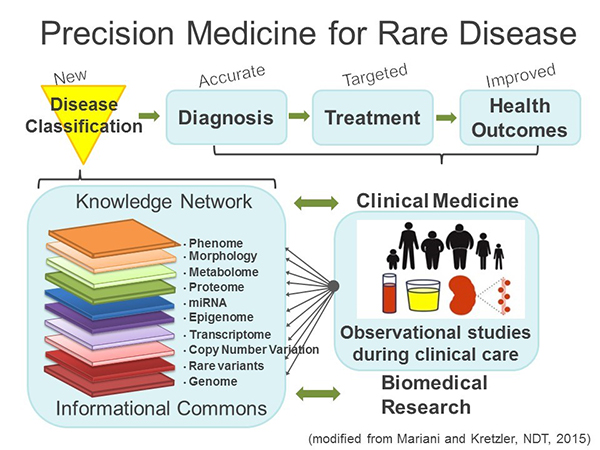
NEPTUNE recruits and prospectively characterizes adults and children with primary glomerular diseases to generate a comprehensive Knowledge Network for nephrotic syndrome. NEPTUNE sites enroll participants at the time of biopsy, and follows them every 4-6 months for a minimum of 36 months, capturing detailed health status data and biofluids at each visit. Using the kidney tissue and biofluids, the consortium has generated deep molecular phenotypes, including individual catalogues of whole genome sequence variation 4, 5, whole genome glomerular and tubulointerstitial transcriptomes assayed by chip 6, 7 and RNAseq, urine and serum cytokine panels assayed using Luminex technology 8, and urinary exosome proteomes 9. NEPTUNE uses the NEPTUNE Digital Pathology Protocol and Scoring System to collect and review kidney biopsy whole slide imaging (WSI), immunofluorescence (IF) and electron microscopy (EM) digital images10-12 which are deposited into the digital pathology repository at the National Cancer Institute. WSI are manually annotated for enumeration of glomeruli at all biopsy levels and the glomerular, tubulointerstitial, and vascular compartments in each biopsy are scored using 84 agnostic and reproducible “descriptors,” many more than are used in conventional classification systems.13 This new scoring system has allowed the study of the predictive value of discrete structural changes that would otherwise escape the analysis when utilizing conventional classification systems, including global sclerosis, interstitial fibrosis and morphologic measurements.14-16 Other elements in the Knowledge Network include data from interviews at each study visit, yielding extensive demographic and socioeconomic status data, clinical data including birth history, personal and family medical history, hospitalizations, body mass index, blood pressure, edema assessment, end-stage kidney disease, and medications. Disease status is derived from centrally measured serum creatinine and urine protein. Patient Reported Outcomes are captured with the PROMIS® Short Forms for adults >18, self-report for children age 8-17 and parent proxy for age 5-10. Census tract is recorded and linked to databases providing a range of social, economic, and environmental conditions. The multiscalar data are then deposited in a secure cloud-based instance of tranSMART, an open-source, web-based systems biology analysis and visualization platform 17, 18 for use by investigators both internal and external to the consortium through its ancillary and publication programs. To-date, the consortium has approved nearly 120 ancillary studies using the NEPTUNE datasets. Key publications that demonstrate the dividends paid by the NEPTUNE precision medicine strategy include: 1. Development of novel outcome measures now adopted for FDA registration in new clinical trials 19; 2. Use of a tissue-transcriptome driven classification of nephrotic syndrome patients to identify a high risk group of patients with tumor necrosis factor (TNF) activation 20; 3. Generation of a pipeline to identify a non-invasive marker panel to measure therapeutic target engagement pathway activity assessment; 7, 21-23 and 4. Support of two recently completed Phase 2 Trials in Nephrotic Syndrome: Randomized, Double-Blind, Safety and Efficacy Study of RE-021 (Sparsentan) in Focal Segmental Glomerulosclerosis (Trachtman 2018 PMID:30361325) and MEmbranous Nephropathy Trial Of Rituximab (Fervenza Nephron). In the next funding cycle NEPTUNE will test the molecular stratification for its power to increase patient responses to targeted treatments currently developed in public private partnerships with NEPTUNE.
Literature Cited
- Schieppati, A, Mosconi, L, Perna, A, Mecca, G, Bertani, T, Garattini, S, Remuzzi, G: Prognosis of untreated patients with idiopathic membranous nephropathy. N Engl J Med, 329: 85-89, 1993.
- Bose, B, Cattran, D, Toronto Glomerulonephritis, R: Glomerular diseases: FSGS. Clin J Am Soc Nephrol, 9: 626-632, 2014.
- Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease, Washington DC, National Academy of Sciences., 2011.
- Debiec, H, Dossier, C, Letouze, E, Gillies, CE, Vivarelli, M, Putler, RK, Ars, E, Jacqz-Aigrain, E, Elie, V, Colucci, M, Debette, S, Amouyel, P, Elalaoui, SC, Sefiani, A, Dubois, V, Simon, T, Kretzler, M, Ballarin, J, Emma, F, Sampson, MG, Deschenes, G, Ronco, P: Transethnic, Genome-Wide Analysis Reveals Immune-Related Risk Alleles and Phenotypic Correlates in Pediatric Steroid-Sensitive Nephrotic Syndrome. J Am Soc Nephrol, 29: 2000-2013, 2018.
- Gillies, CE, Putler, R, Menon, R, Otto, E, Yasutake, K, Nair, V, Hoover, P, Lieb, D, Li, S, Eddy, S, Fermin, D, McNulty, MT, Nephrotic Syndrome Study, N, Hacohen, N, Kiryluk, K, Kretzler, M, Wen, X, Sampson, MG: An eQTL Landscape of Kidney Tissue in Human Nephrotic Syndrome. Am J Hum Genet, 103: 232-244, 2018.
- Sampson, MG, Robertson, CC, Martini, S, Mariani, LH, Lemley, KV, Gillies, CE, Otto, EA, Kopp, JB, Randolph, A, Vega-Warner, V, Eichinger, F, Nair, V, Gipson, DS, Cattran, DC, Johnstone, DB, O'Toole, JF, Bagnasco, SM, Song, PX, Barisoni, L, Troost, JP, Kretzler, M, Sedor, JR, Nephrotic Syndrome Study, N: Integrative Genomics Identifies Novel Associations with APOL1 Risk Genotypes in Black NEPTUNE Subjects. J Am Soc Nephrol, 27: 814-823, 2016.
- Ju, W, Nair, V, Smith, S, Zhu, L, Shedden, K, Song, PX, Mariani, LH, Eichinger, FH, Berthier, CC, Randolph, A, Lai, JY, Zhou, Y, Hawkins, JJ, Bitzer, M, Sampson, MG, Thier, M, Solier, C, Duran-Pacheco, GC, Duchateau-Nguyen, G, Essioux, L, Schott, B, Formentini, I, Magnone, MC, Bobadilla, M, Cohen, CD, Bagnasco, SM, Barisoni, L, Lv, J, Zhang, H, Wang, HY, Brosius, FC, Gadegbeku, CA, Kretzler, M, Ercb, CPN, Consortium, PK-I: Tissue transcriptome-driven identification of epidermal growth factor as a chronic kidney disease biomarker. Sci Transl Med, 7: 316ra193, 2015.
- Mariani, LH, Eddy, S, Martini, S, Eichinger, F, Godfrey, B, Nair, V, Adler, SG, Appel, GB, Athavale, A, Barisoni, L, Brown, E, Cattran, DC, Dell, KM, Derebail, V, Fervenza, FC, Fornoni, A, Gadegbeku, CA, Gibson, KL, Gipson, D, Greenbaum, LA, Hingorani, SR, Hlandunewich, MA, Hogan, J, Holzman, LB, Jefferson, JA, Kaskel, FJ, Kopp, JB, Lafayette, RA, Lemley, KV, Lieske, JC, Lin, J-J, Myers, KE, Nachman, PH, Nast, CC, Neu, AM, Reich, HN, Sambandam, K, Sedor, JR, Sethna, CB, Srivastava, T, Trachtman, H, Tran, C, Wang, C-s, Kretzler, M: Redefining Nephrotic Syndrome in Molecular Terms: Outcome-associated molecular clusters and patient stratification with noninvasive surrogate biomarkers. bioRxiv, 2018.
- Hogan, MC, Johnson, KL, Zenka, RM, Charlesworth, MC, Madden, BJ, Mahoney, DW, Oberg, AL, Huang, BQ, Leontovich, AA, Nesbitt, LL, Bakeberg, JL, McCormick, DJ, Bergen, HR, Ward, CJ: Subfractionation, characterization, and in-depth proteomic analysis of glomerular membrane vesicles in human urine. Kidney Int, 85: 1225-1237, 2014.
- Barisoni L, Nast CC, Jennette JC, et al. Digital pathology evaluation in the multicenter Nephrotic Syndrome Study Network (NEPTUNE). Clin J Am Soc Nephrol. 2013;8(8):1449-1459.
- Barisoni L, Troost JP, Nast C, et al. Reproducibility of the NEPTUNE descriptor-based scoring system on whole-slide images and histologic and ultrastructural digital images. Mod Pathol. 2016;29(7):671-684.
- Zee J, Hodgin JB, Mariani LH, et al. Reproducibility and Feasibility of Strategies for Morphologic Assessment of Renal Biopsies Using the Nephrotic Syndrome Study Network Digital Pathology Scoring System. Arch Pathol Lab Med. 2018;142(5):613-625.
- Rosenberg AZ, Palmer M, Merlino L, et al. The Application of Digital Pathology to Improve Accuracy in Glomerular Enumeration in Renal Biopsies. PLoS One. 2016;11(6):e0156441.
- Lemley KV, Bagnasco SM, Nast CC, et al. Morphometry Predicts Early GFR Change in Primary Proteinuric Glomerulopathies: A Longitudinal Cohort Study Using Generalized Estimating Equations. PLoS One. 2016;11(6):e0157148.
- Mariani LH, Martini S, Barisoni L, et al. Interstitial fibrosis scored on whole-slide digital imaging of kidney biopsies is a predictor of outcome in proteinuric glomerulopathies. Nephrol Dial Transplant. 2018;33(2):310-318.
- Hommos MS, Zeng C, Liu Z, et al. Global glomerulosclerosis with nephrotic syndrome; the clinical importance of age adjustment. Kidney Int. 2018;93(5):1175-1182.
- Scheufele, E, Aronzon, D, Coopersmith, R, McDuffie, MT, Kapoor, M, Uhrich, CA, Avitabile, JE, Liu, J, Housman, D, Palchuk, MB: tranSMART: An Open Source Knowledge Management and High Content Data Analytics Platform. AMIA Jt Summits Transl Sci Proc, 2014: 96-101, 2014.
- Athey, BD, Braxenthaler, M, Haas, M, Guo, Y: tranSMART: An Open Source and Community-Driven Informatics and Data Sharing Platform for Clinical and Translational Research. AMIA Jt Summits Transl Sci Proc, 2013: 6-8, 2013.
- Troost, JP, Trachtman, H, Nachman, PH, Kretzler, M, Spino, C, Komers, R, Tuller, S, Perumal, K, Massengill, SF, Kamil, ES, Oh, G, Selewski, DT, Gipson, P, Gipson, DS: An Outcomes-Based Definition of Proteinuria Remission in Focal Segmental Glomerulosclerosis. Clin J Am Soc Nephrol, 13: 414-421, 2018.
- Mariani, LH, Martini, S, Barisoni, L, Canetta, PA, Troost, JP, Hodgin, JB, Palmer, M, Rosenberg, AZ, Lemley, KV, Chien, HP, Zee, J, Smith, A, Appel, GB, Trachtman, H, Hewitt, SM, Kretzler, M, Bagnasco, SM: Interstitial fibrosis scored on whole-slide digital imaging of kidney biopsies is a predictor of outcome in proteinuric glomerulopathies. Nephrol Dial Transplant, 33: 310-318, 2018.
- Eddy, S, Nair, V, Mariani, LH, Eichinger, FE, Hartman, J, Huang, H, Parikh, H, Taroni, JN, Lindenmeyer, MT, Ju, W, Greene, CS, Grayson, PC, Godfrey, B, Cohen, CD, Sampson, MG, Lafayette, RA, Krischer, J, Merkel, PA, Kretzler, M: Inflammatory and JAK-STAT Pathways as Shared Molecular Targets for ANCA-Associated Vasculitis and Nephrotic Syndrome. bioRxiv, 2018.
- Grayson, PC, Eddy, S, Taroni, JN, Lightfoot, YL, Mariani, L, Parikh, H, Lindenmeyer, MT, Ju, W, Greene, CS, Godfrey, B, Cohen, CD, Krischer, J, Kretzler, M, Merkel, PA, Vasculitis Clinical Research Consortium, tERcBc, the Nephrotic Syndrome Study, N: Metabolic pathways and immunometabolism in rare kidney diseases. Ann Rheum Dis, 77: 1226-1233, 2018.
- Eddy, S, Nair, V, Mariani, LH, Eichinger, FE, Hartman, J, Huang, H, Parikh, H, Taroni, JN, Lindenmeyer, MT, Ju, W, Greene, CS, Grayson, PC, Godfrey, B, Cohen, CD, Sampson, MG, Lafayette, RA, Krischer, J, Merkel, PA, Kretzler, M: Inflammatory and JAK-STAT Pathways as Shared Molecular Targets for ANCA-Associated Vasculitis and Nephrotic Syndrome. bioRxiv, 2018.


